Scalable learning for bridging the species gap in image-based plant phenotyping
This is the webpage for the (under review) CVIU paper: Scalable learning for bridging the species gap in image-based plant phenotyping
[ Paper ] [ BibTex ] [ Dataset (coming soon - here is our previous dataset] [ Pretrained Model (coming soon - here is our previous pretrained model)]
Abstract
Our synthetic dataset and pretrained model are available at https://bitbucket.csiro.au/scm/ag3d/leaf_segmenter_public.git.
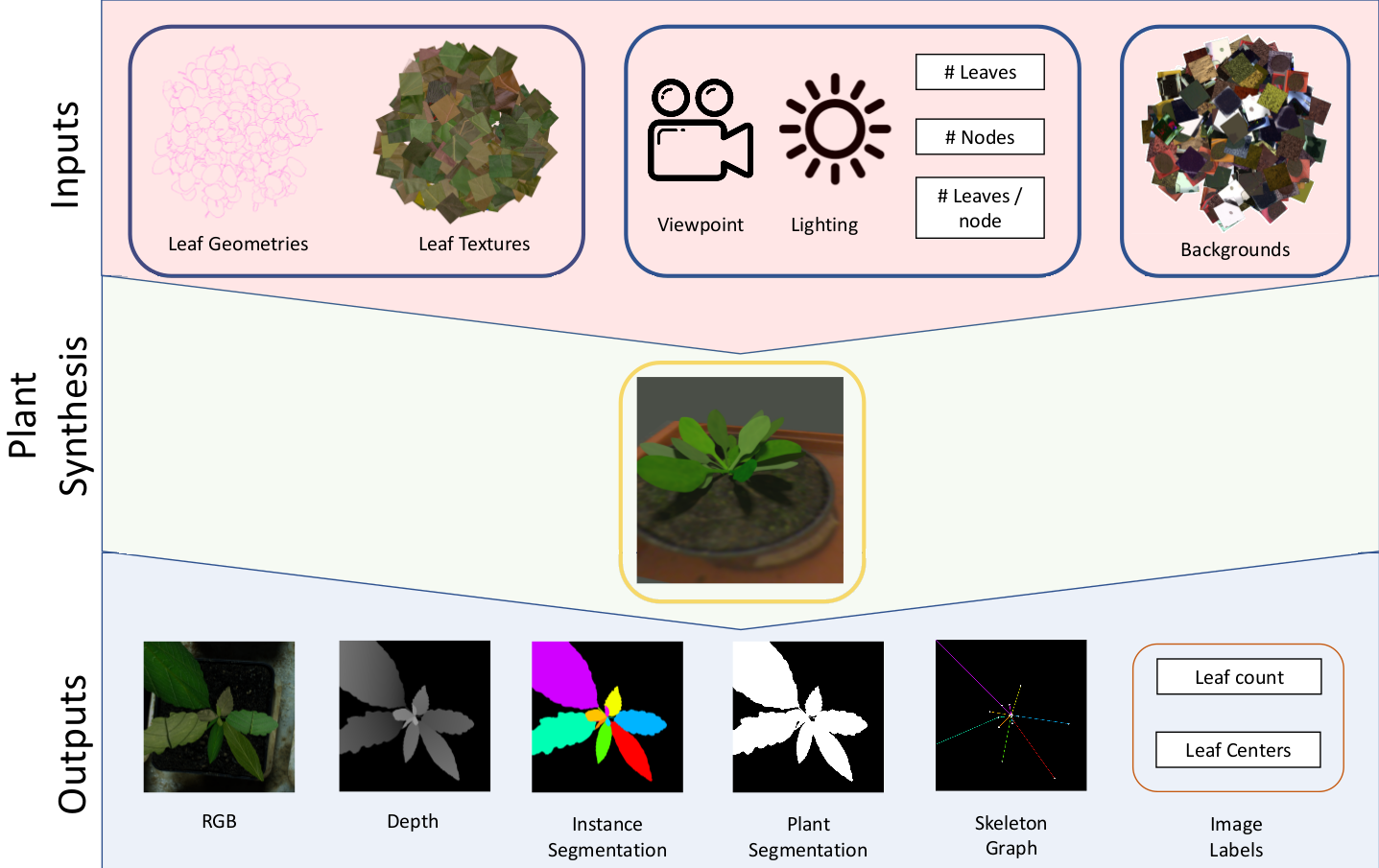
Synthetic Data Generation
We present, UPGen, a simulation based data pipeline which produces annotated synthetic images of plants. Our approach leverages Domain Randomisation (DR) concepts to model stochastic biological variation between plants of the same and different species. Training plant image analysis algorithms on our data learns a model which is robust to domain and species.
Our method has several benefits over other synthetic data approaches such as the use of GANs. These include:
- Fine control of data parameters - You have control over the dataset distribution.
- Fine control of the pipeline inputs - Easily swap in and out textures and leaf geometries to match your application domain to boost performance.
Example synthetic images and the corresponding leaf instance segmentation masks.
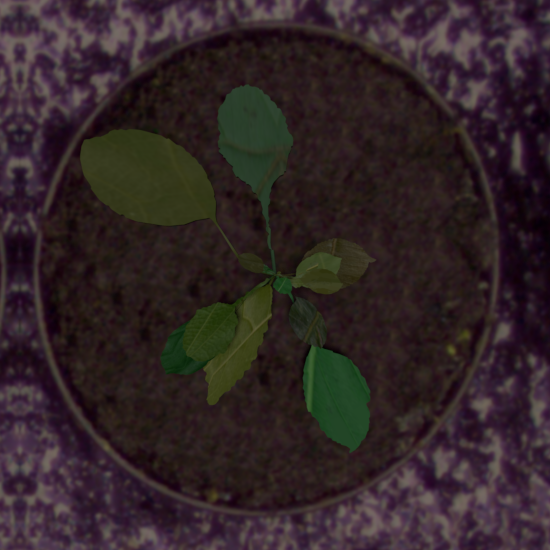



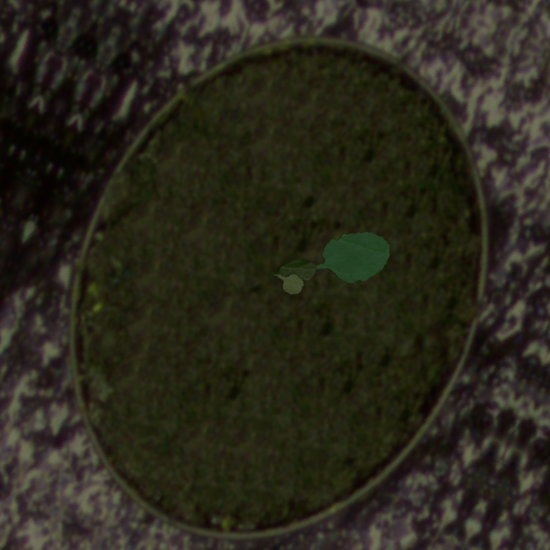

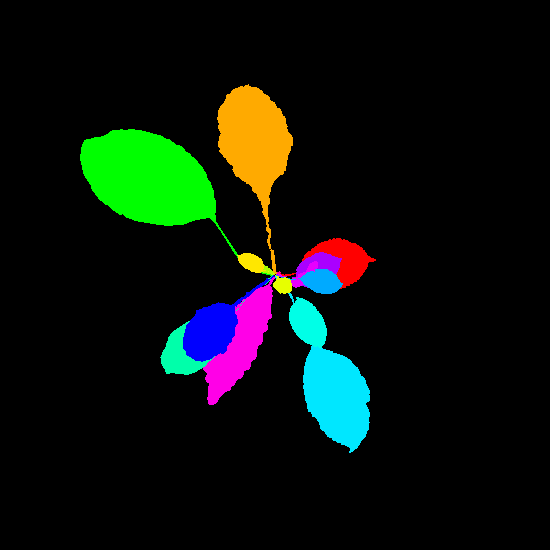
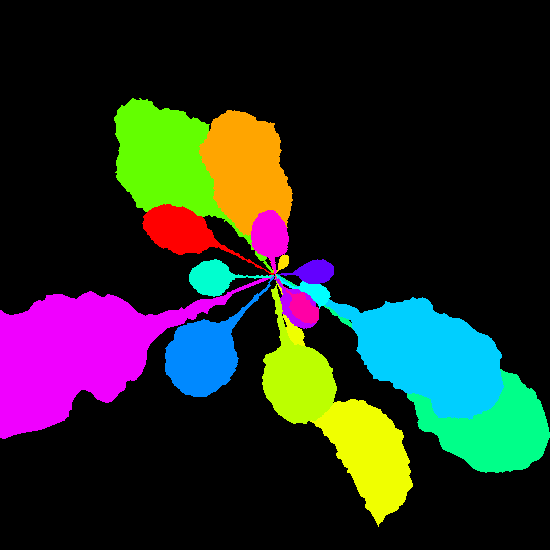
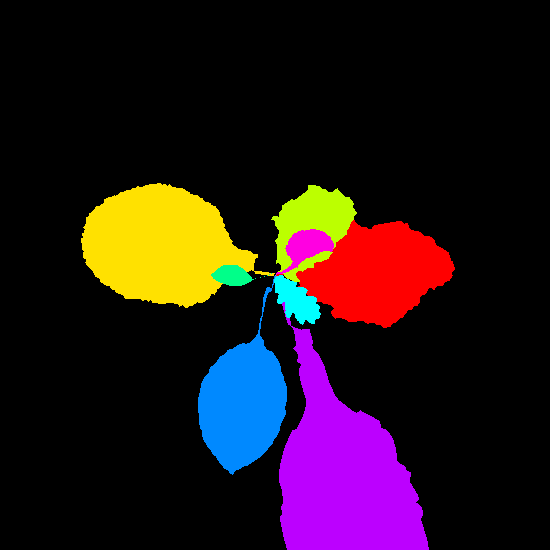

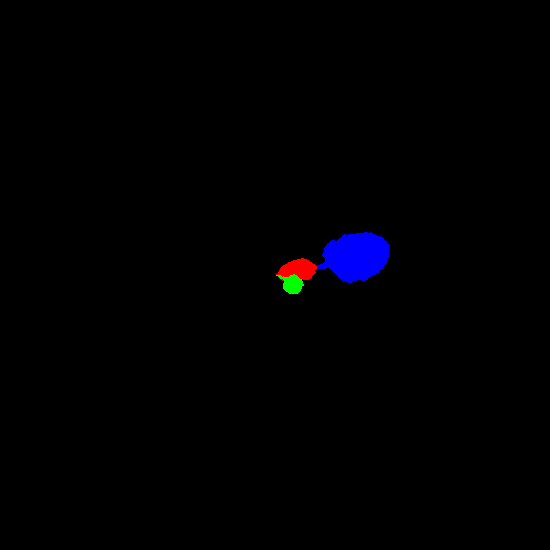
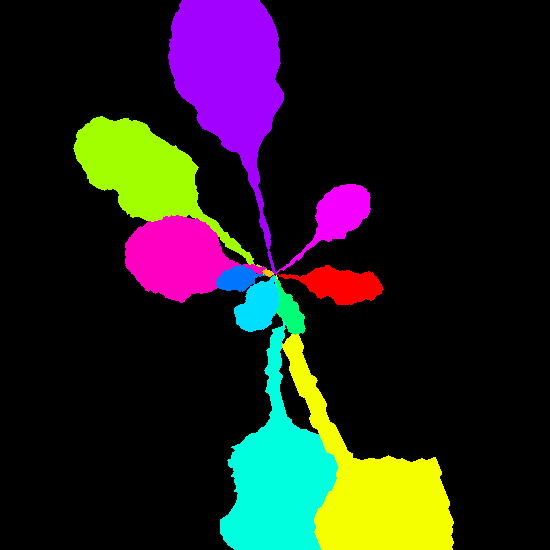
Dataset
Our dataset will be available soon. Until then, our previous dataset is available for download here
Results
We validate our approach on the task of leaf instance segmentation.
In the wild
We train Mask-RCNN using our data and evauluate it on two completely unseen plant datasets, the Komatsuna Dataset and an in-house capsicum dataset. Using our data for training significantly outperformed the baseline of training on publicly available plant datasets.
Example segmentations on the Komatsuna dataset (left) and the capsicum dataset (right).


The CVPPP Leaf Segmentation Challenge
We also compete in the Computer Vision Problems in Plant Phenotyping (CVPPP) Leaf Segmentation Challenge (LSC). We achieve state of the art performance training Mask-RCNN using our synthetic data.
Example segmentations on test images. Left to right is an image from each test dataset: A1, A2, A3 and A4.




Citation
@article{ward2020scalable,
title={Scalable learning for bridging the species gap in image-based plant phenotyping},
author={Ward, Daniel and Moghadam, Peyman},
journal={arXiv preprint arXiv:2003.10757},
year={2020}
}